Research Overview
The research interests of the Kumar laboratory are centered on the (1) use of chemistry to design molecules to interrogate and illuminate fundamental mechanisms in biology, or be used as therapeutics; and (2) use of biology to “evolve” and “select” molecules that can perform chemistry in non-biological and medicinal settings.
These are some questions we are trying to answer: (i) Is it possible to design and mimic natural proteins and other biological macromolecules by use of building blocks that nature does not use – and whether such constructs can be endowed with properties that are not found in biology?; (ii) How did the first enzymes arise in the imagined Darwin’s pond – is there a way to recreate this scenario and in the process develop a fundamentally new method to create enzymes?; (iii) Biology uses phase separation, that is, clustering of different compounds in confined locations – a process that is key in orchestrating the daily activities of a cell – can we find methods that can predictably dictate where molecules are located in a given environment and thereby direct the phenotype that is generated?; (iv) Can we rationally design small molecules and peptides that can function against antibiotic resistant bacteria that are threatening the most basic tenet of modern medicine?
Chemical Building Blocks not Found in Nature

The accelerated sequencing of whole organism genomes and their analysis poses a tremendous challenge to biochemists: the attendant elucidation of the structure and function of the corresponding proteins that are encoded.
These efforts will no doubt reveal the functions of individual protein molecules, many of which are likely to be novel and unprecedented. For thirty years protein scientists have been studying the relationship between protein sequence, structure and function by constructing mutants of known proteins. By comparison, organic chemists have spent two centuries developing reactions, of which many are biologically important but many more are unknown in living organisms. Many of the important technological innovations have come from reactions that are not currently known to exist in nature’s arsenal. Inspired by such success, protein scientists have begun to ask ‘Rather than limit our vision to the set of extant proteins, can we rationally design and construct novel structures and endow them with predetermined and unusual functions?’.
Our laboratory has designed several proteins that are “non-stick”, very much like Teflon®, and have a variety of useful properties, as probes in biology and as therapeutics as antibiotics that are stable. In addition, we have taken this idea further and have used it to coat cancer cells with a Teflon® like material that is currently being tested for use in preventing metastasis, and in imaging tumors by 19F MRI before they are visible by other means. Related efforts have recently led to the uncovering of structural features of receptors on cell surfaces that are mediators of signaling, and have resulted in the design of molecules that perform better than the best selling current drugs for diabetes and short bowel syndrome.
Fluorinated Cells:
- Fluorogenic Sialic Acid Glycosides for Quantification of Sialidase Activity upon Unnatural Substrates. Bioorg. Med. Chem. Lett. 2013, 23 (11), 3406–3410. [PubMed]
- Modulation of Cellular Adhesion by Glycoengineering. J. Med. Chem. 2010, 53 (10), 4277–4284. [PubMed]
- Fluorination of Mammalian Cell Surfaces via the Sialic Acid Biosynthetic Pathway. Bioorg. Med. Chem. Lett. 2008, 18 (22), 5945–5947. [PubMed]
Fluorinated Peptides:
- Biosynthesis and Stability of Coiled-Coil Peptides Containing (2S,4R)-5,5,5-Trifluoroleucine and (2S,4S)-5,5,5-Trifluoroleucine. ChemBioChem 2008, 10 (1), 84–86. [PubMed]
- Programmed Self-Sorting of Coiled Coils with Leucine and Hexafluoroleucine Cores. Bilgiçer, B.; Xing, X.; Kumar, K. J. Am. Chem. Soc. 2001, 123 (47), 11815–11816. [PubMed]
Novel Antibiotics

The most basic tenet of modern medicine is being threatened by drug resistant bacteria. Antibiotics have saved millions of lives but the emergence of resistant strains has resulted in only two “last-resort” drugs that are left to treat patients infected with multiple drug resistant bacteria. We have started a program that will result in the “selection” of peptides that are right-handed (this is opposite of the handedness seen in normal natural proteins), therefore making them stable as they cannot be degraded by standard metabolic pathways. This project utilizing evolution and selection cycles using ‘phage display’, a technique that uses manipulation of a viral genome to rapidly screen hundreds of millions of compounds.
These projects represent an ambitious agenda of using chemistry to understand biological processes and develop therapeutics; and of using biology to provide molecules that will serve as chemical agents in reactions and in medicinal settings.
Selected Publication:
- Discovery of Peptide Antibiotics Composed of D-Amino Acids. ACS Chem. Biol. 2019, 14, 1498-1506. [PubMed]
- Antimicrobial Activity and Protease Stability of Peptides Containing Fluorinated Amino Acids. J. Am. Chem. Soc. 2007, 129 (50), 15615–15622. [PubMed]
Visualizing Compartmentalization in Biology
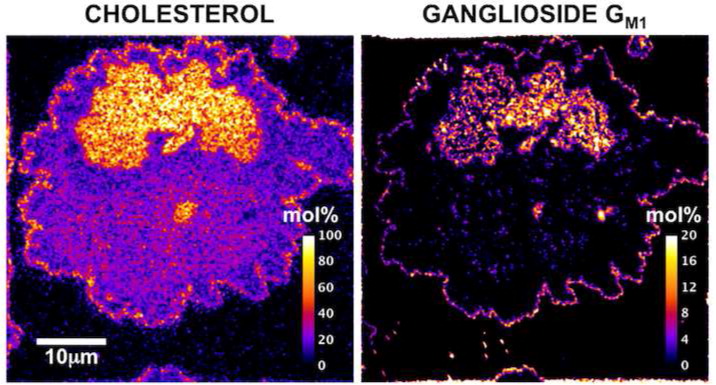
Biology compartmentalizes molecules in various places inside the cell and in the plasma membrane that envelops the cell. Assemblies containing special lipids and proteins, called “lipid rafts” have been invoked, and these play a vital role in various processes from signaling of messages, to cancer metastasis. However, these “rafts” are small and dynamic and it has proven difficult to visualize such ephemeral entities. We have recently shown the existence of a combined phase of a lipid isolated from bovine brain and cholesterol postulated to form a compartmentalized assembly within the plasma membrane using 2-D mass spectrometry. We are now extending these studies in living cells. The ability to present molecules on the surface of a living cell that are clustered versus individual isolated floating compounds may be helpful in revealing fundamental mechanisms in biology.
Selected Publications:
- Colocalization of the Ganglioside GM1 and Cholesterol Detected by Secondary Ion Mass Spectrometry. J. Am. Chem. Soc. 2013, 135 (15), 5620–5630. [PubMed]
- Trifluoromethyl Derivatization of the Ganglioside, GM1. Synthesis. 2010, 1905–1908.
- Fluorinated Lipid Constructs Permit Facile Passage of Molecular Cargo into Living Cells. J. Am. Chem. Soc. 2009, 131 (34), 12091–12093. [PubMed]
Enzyme Evolution

The evolution of proteins by random assembly of amino acids to result in both form and function is an unlikely process. The sheer probability of a single enzyme containing 250 amino acid residues to be formed randomly (1/20250) is enough to exhaust all time and carbon in the universe before all possibilities of a single protein have been made many times over. Indeed, it has been suggested that the likelihood of a hurricane blowing through a junkyard and assembling a Boeing 747 is enormously larger than assembling a functional enzyme from random components. There have been other analogies of a similar kind but the sheer magnitude of the possibilities leaves us with the question of how did modern day enzymes evolve to catalyze reactions? Our research program is using clues from the structure of modern day and ancient enzymes to “dissect” them, and learn to evolve new chemical function from the extant set of proteins. This results in proteins that are built from blocks that are 30-40 residues long and these have a high probability of having existed in Darwin’s imagined pond early in evolution.
